Insight into power generation in photosynthesis may lead to more resilient crops
A study into the energy-making process in plants could help engineer crops more resistant to stress or bacteria that produce pharmaceuticals.
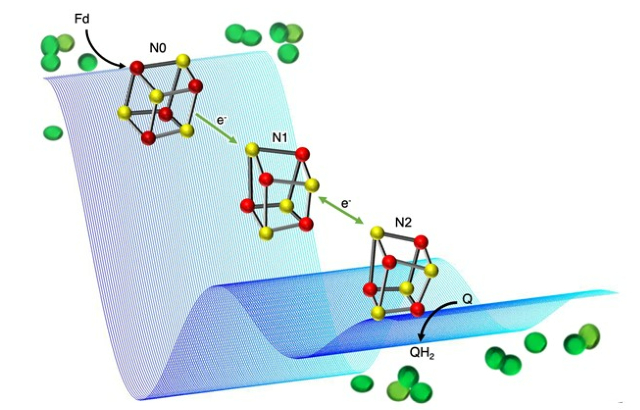
Relay of iron-sulfur clusters (red-yellow cubanes) that wire electrons from ferredoxin (Fd) to plastoquinone (Q) in photosynthetic complex I from cyanobacteria (schematically shown in green). Example double electron-electron resonance (DEER) traces are shown in blue – DEER is the pulse EPR technique used to assign the properties of the clusters to their location in the protein structure.
A team of researchers, led by Imperial College London and Queen Mary University of London, have mapped a key process in energy conversion in plants.
The study, published today in Nature Communications, could help scientists improve the resilience of important crops and engineer bacteria that can produce useful compounds like pharmaceuticals more sustainably.
All cells, whether plant or animal, use a chemical called adenosine triphosphate (ATP) as their energy currency. Animals like humans make ATP through the process of respiration, whereas plants use photosynthesis to convert sunlight into ATP.
There is an intermediate step in the process in plant cells, where the light energy is used to create a ‘proton gradient’, which then helps create ATP. The proton gradient is like the height of the water in a hydroelectric dam – it is a form of ‘potential energy’, which then gets converted to the chemical energy unit of ATP.
However, how the proton gradient is created in the first place is poorly understood. This is particularly true for one part of the molecular machinery plants use in this process, called photosynthetic complex 1 (PS-C1).
Mapping electron transfer
Researchers know that PS-C1 is used by plants, including many important crop plants, to increase ATP production at times of high stress, helping them continue to grow under difficult conditions such as low light, drought, or high temperatures.
Now, the team behind the new study have mapped how an electron transfer process required for setting up a proton gradient in PS-C1 works, providing key information about how plants and photosynthetic bacteria can gain extra energy.
Lead researcher Dr Maxie Roessler, from the Department of Chemistry at Imperial, said: “With this study we contribute to understanding how cells convert the potential energy in proton gradients into chemical energy – a process which underpins nearly all life on earth.”
Co-lead Dr Guy Hanke, from Queen Mary, added: “Ultimately, we hope that understanding how PS-C1 works will enable us, and other researchers, to manipulate photosynthetic bacteria for biotechnological purposes. This could include supporting the energetic demands necessary to produce high-value compounds such as pharmaceuticals in a more sustainable way.”
Improving our understanding
The research team hopes that a better understanding of PS-C1 will ultimately help to develop crops that can better withstand the stresses induced by climate change, such as temperature fluctuations and drought.
PS-C1 is embedded in the membrane of plant chloroplasts, allowing it to control the movement of molecules across the membrane. The researchers knew that PS-C1 must act as a ‘proton pump’ to create the proton gradient across the membrane, but how it does this has been difficult to study, as PS-C1 is such a large and complex molecular machine.
To study PS-C1, the team used a technique called electron paramagnetic resonance (EPR) spectroscopy to map the position of single (unpaired) electrons. The transfer of electrons between parts of PS-C1 powers the movement of protons, so mapping the electron transfer path helps to provide insight into the way protons move across the membrane.
The team hope this fundamental study will open the door for greater understanding of how this molecular machine works, and of potential ways to harness its ability to create extra energy resources for plants and bacteria that use photosynthesis.
More information
- Research publication: ‘Functional basis of electron transport within photosynthetic complex I’ by Katherine Richardson et al. is published in Nature Communications.